Annotation
����������� We developed a controlled vocabulary to annotate gene expression patterns during embryogenesis in collaboration with Volker Hartnestein and Michael Ashburner. The basic concept of annotating gene expression patterns during developmental process is the requirement for a unique term for an embryonic structure at each stage of development. In the context of Drosophila embryogenesis we need to be able to name not only the differentiated embryonic structure, but also all the developmental intermediates that lead to its formation. As far as de novo gene expression is concerned Drosophila embryogenesis can be described as a process that turns a simple epithelial monolayer represented by cellular blastoderm, by a series of cell proliferations, differentiations and movements, into a highly complex embryo with all major organ system formed.
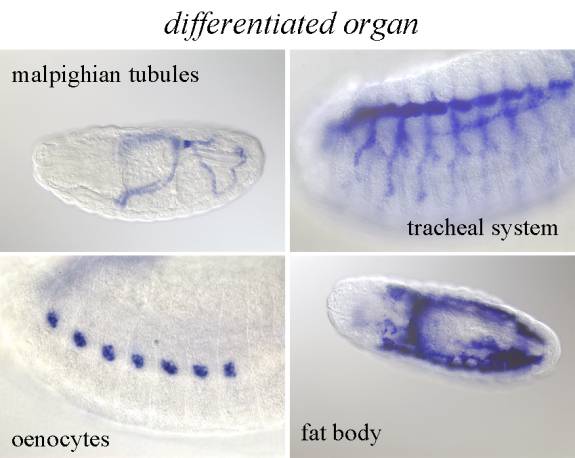
����������� Differentiated organs: At one end of the above-described process of embryogenesis are the differentiated organs that can be distinguished based on their unique morphology and usually have a well-established name. We use anatomical terms from Flybase controlled vocabulary for anatomy without any suffix to annotate those structures.
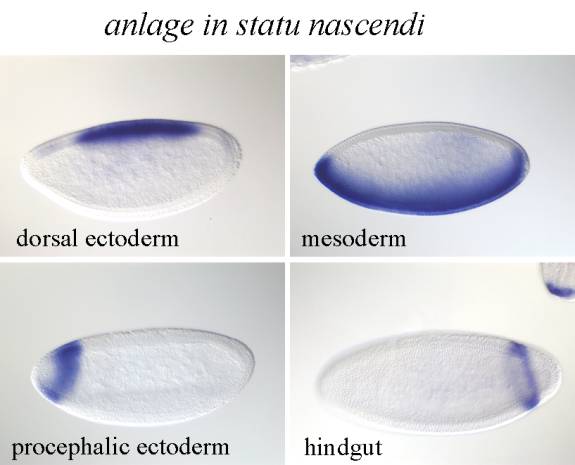
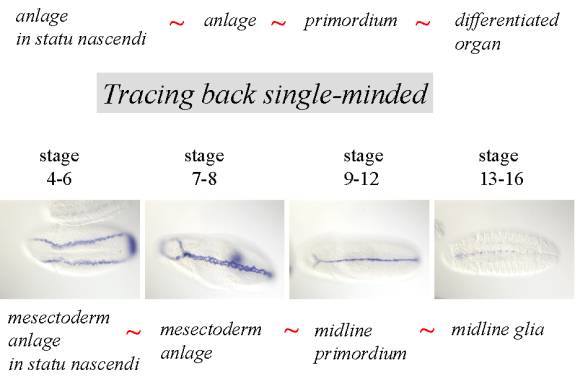
����������� Anlage in statu nascendi : Gene expression patterns can be first observed at the cellular blastoderm stage when zygotic gene expression begins. Volker Hartenstein introduced a concept of anlage in statu nascendi (loosely translated as anlage in the process of being formed) that is defined as a morphologically indistinct group of cells at cellular blastoderm that can be distinguished only on the basis of gene expression and does not correspond in its extend to any single later appearing structure. We subdivide the cellular blastoderm epithelium so that each cell can be assigned to a anlage in statu nascendi.
����������� We distinguish two types of developmental intermediates
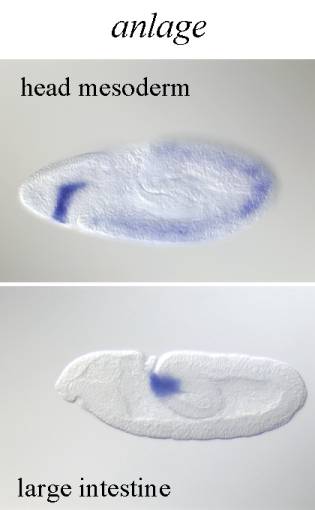
����������� Anlage: Similarly to anlage in statu nascendi anlage is morphologically indistinct defined only by gene expression however it already does correspond in its extent to the organ or primordium that develops from it. Generally developmental structures will be called anlage later in development after the cellular blastoderm stage. Anlage usually gives rise to a primordium however not every primordium necessarily has a corresponding anlage.�
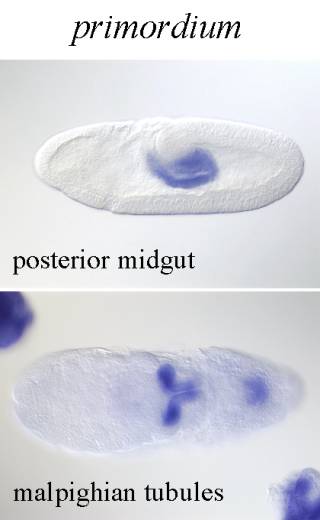
����������� Primordium : Primordium is similar to differentiated organ in that it can be identified based on its unique morphology. Primordium usually develops from an anlage, and can give rise to one or more differentiated organs. If within a primordium a group of morphologically indistinct cells that show expression of specific gene is found, such a group will be named a specific anlage. It is part of a primordium and gives rise to subset of organs that develop from that primordium or to a subset of the differentiated organ.
 ����
���� 
����������� Each annotation term is tied to a stage-range. Traditionally embryogenesis has been divided into discreet stages based on morphological landmarks. For practical reasons since it is time consuming to precisely stage every photographed embryo we decided to group embryogenesis stages into six specific stage-ranges. The rational for defining border between the stage-ranges is that at certain times of development such as gastrulation or the onset of organogenesis changes in gene expression are more likely and therefore there is a requirement for a unique naming of the emerging embryonic structures. As discussed in the section about imaging, gene expression patterns are documented collectively by a group of images tied to a specific stage-range. Similarly annotation terms are associated with a unique stage-range and during annotation a subset of the available named structures is linked to the evidence images. Like that group of annotation terms that is documented by group of images at each stage-range constitutes the first level of annotation that can be search computationally and refined by inspection of the raw image data by a motivated expert user.
����������� The annotation terms are linked to one another by relationships that reflect mainly the way they develop from one another (develops from type relationship) or the way they encompass each other (part of relationship). Such a scheme of controlled vocabulary terms connected by relationship is best computationally implemented using Gene Ontology. All our annotation terms have been incorporated into the Flybase controlled vocabulary for anatomy, which will allow comparison with expression data assembled from the literature, expansion of annotation to greater detail and sophisticated searches of the dataset based on Gene Ontology database schema.
����������� To demonstrate the utility of the annotation approach we show here expression pattern for gene single-minded that is expressed in midline glia and the origin of the staining can be traced back all the way to cellular blastoderm through an array of named intermediates.
����������� �